Research area
We are going through the way where others did not go . . .Background
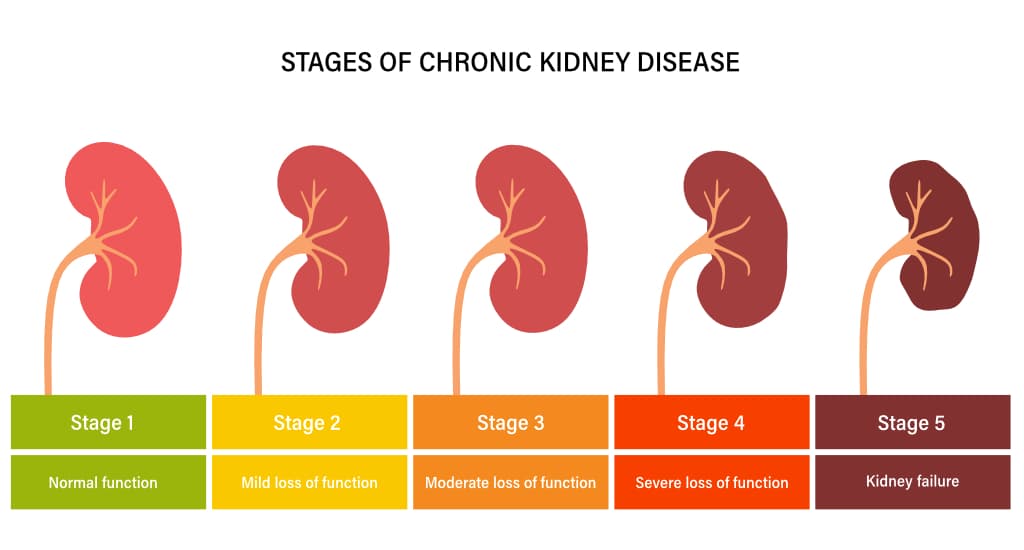
Chronic Kidney Disease (CKD)
is highly prevalent (15%) and mortal disease (109.7 per 1,000 patient-years) [1]. The proposed risk factors of CKD, including age, race, diabetes mellitus, hypertension, and cardiovascular disease, explain only a small fraction of it [2]. Such limited explainability of those risk factors restricts physicians in identifying patients who will experience rapid estimated glomerular filtration rate (eGFR) decline and progress to end-stage renal disease (ESRD).Serum uric acid (SUA) and its related purine metabolites
It is the potential to be a biomarker indicating the CKD incidence and its progression. One proposed mechanism of hyperuricemia leading to CKD progression [3] is via activation of the renal renin-angiotensin system [4-8] and inflammation [9, 10]. SUA is a significant predictor of eGFR decline and albuminuria [11, 12] and is related to kidney-related mortality [13, 14]. Therefore, it is critical to examine SUA levels and their relation to kidney health and eventually find an optimal level of the SUA to preserve kidney health and prevent kidney failure.By integrating the prospective cohort studies and quantitative modeling, we will identify uric acid, as an early kidney function decline biomarkers in purine metabolism, with prospective cohort studies and investigate the predictive role of SUA in non-diabetic CKD progression. This model will give an individual prediction to prevent CKD progression in clinical practice.
Urate Dynamicx
We build models exploring the urate dynamics in a purine metabolic pathway,
and we also explores real-world market medicine having a hidden feature controlling the serum
urate
level.
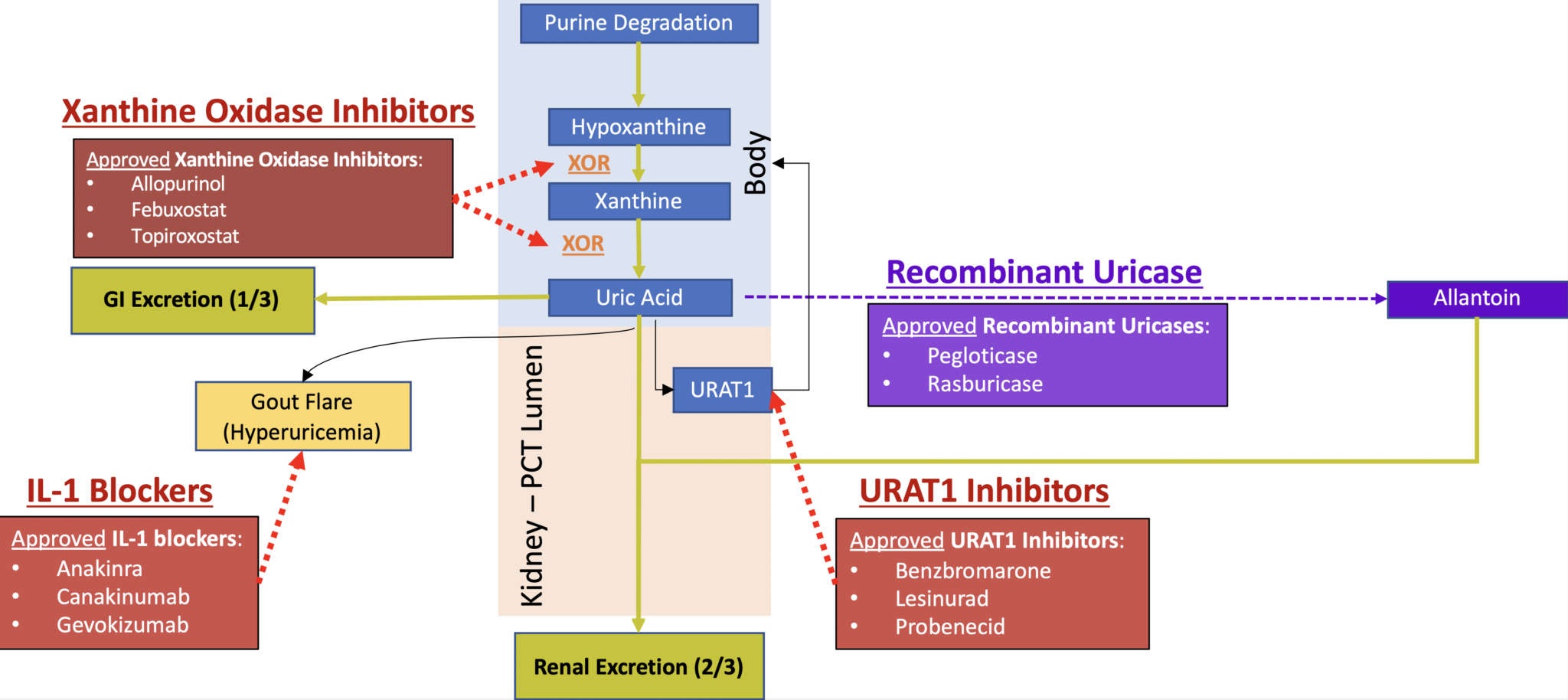
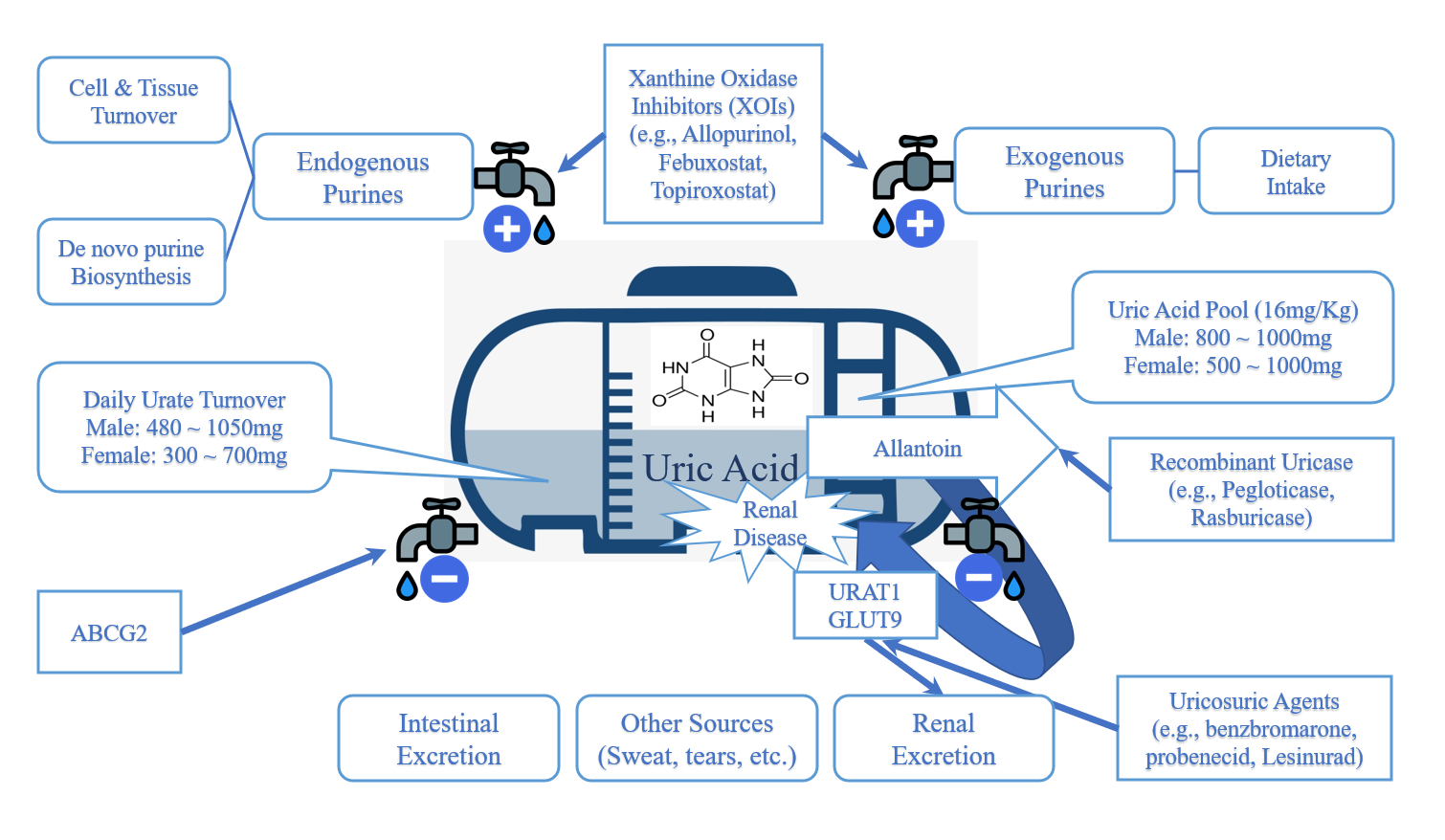
We understand uric acid as an agent
reflecting the dynamic equilibrium of
the purine metabolic pathway.
Genetic Epidemiology & Population Genetics
Our lab identifies uric acid-related genes
and transporters. Each gene is good target for Urate lowering therapy (ULT).
We’re dedicating to develop the next generation ULT! It is coming and new future is near.
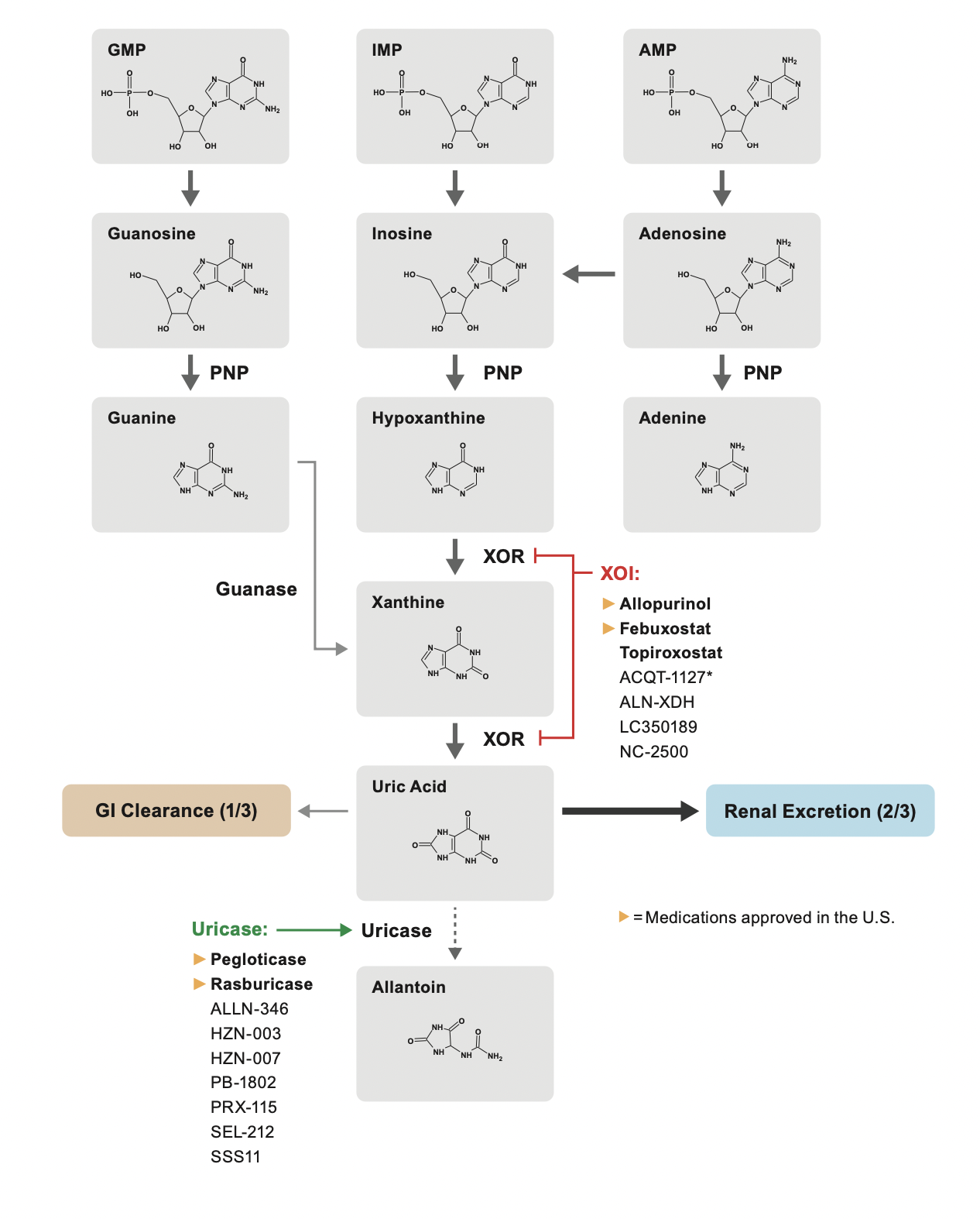
We’re dedicating to develop the next generation ULT! It is coming and new future is near.

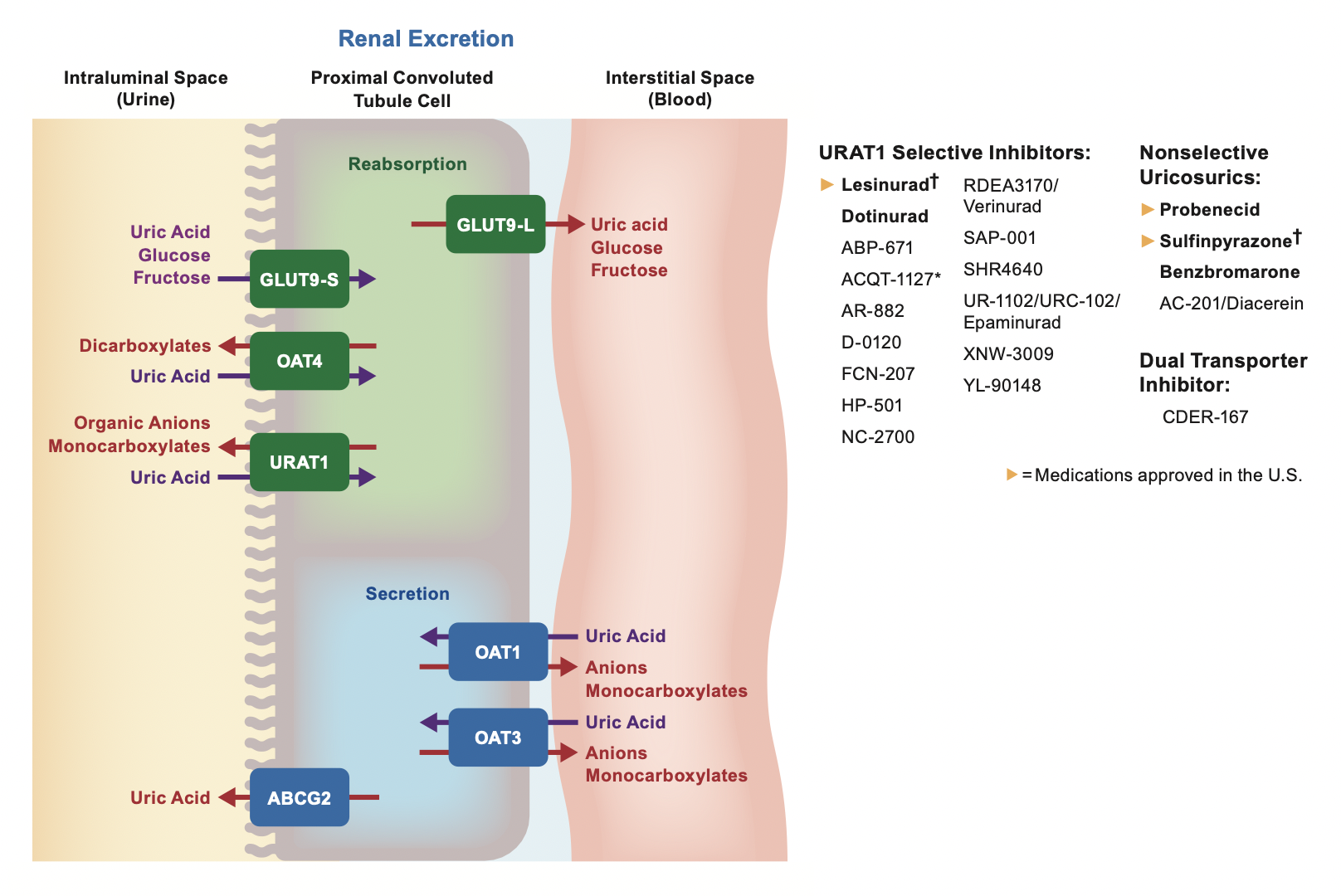
Our goal is to investigate whether
uric acid-related genes and transporters
are related to chronic diseases, especially
chronic kidney disease (CKD)
Clinical Pharmacology
Our lab has contributed to clinical pharmacology and is passionate about new drug development.
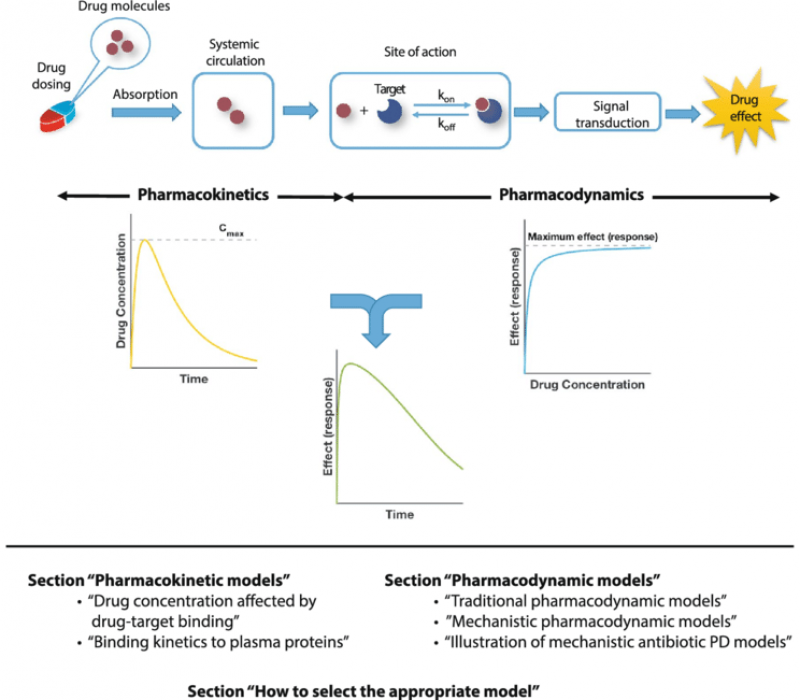
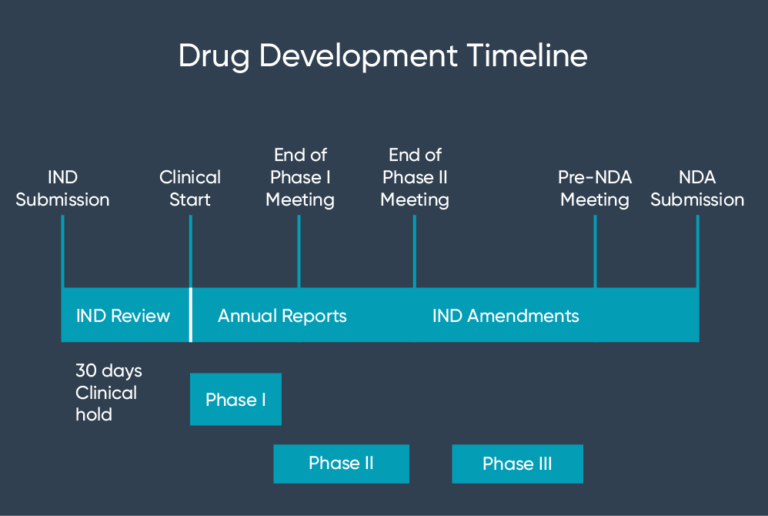
We consult the investigational
new drug (IND) application
and eventually submit a new drug
application (NDA).
Human Data Science
Our lab examines uric acid and its relation to
clinical and risk factors in human health.
We build a prediction model with the data-driven
hypotheses, ultimately identifying not-yet-known
causal factors for chronic diseases.
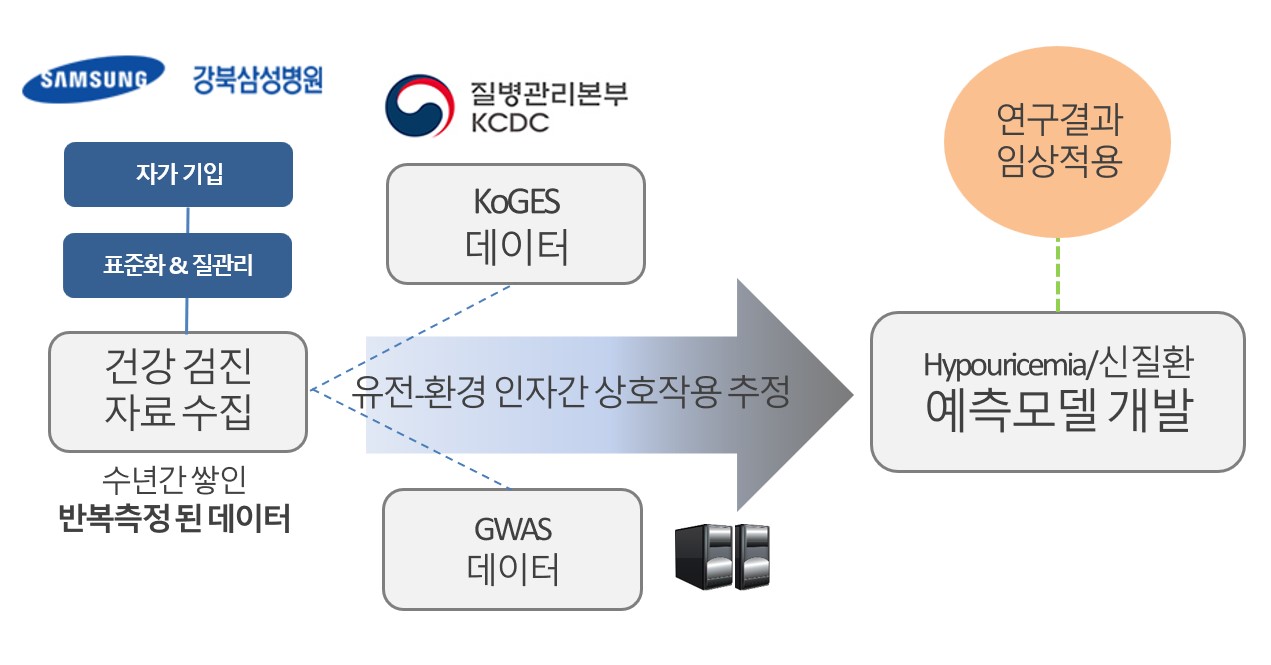
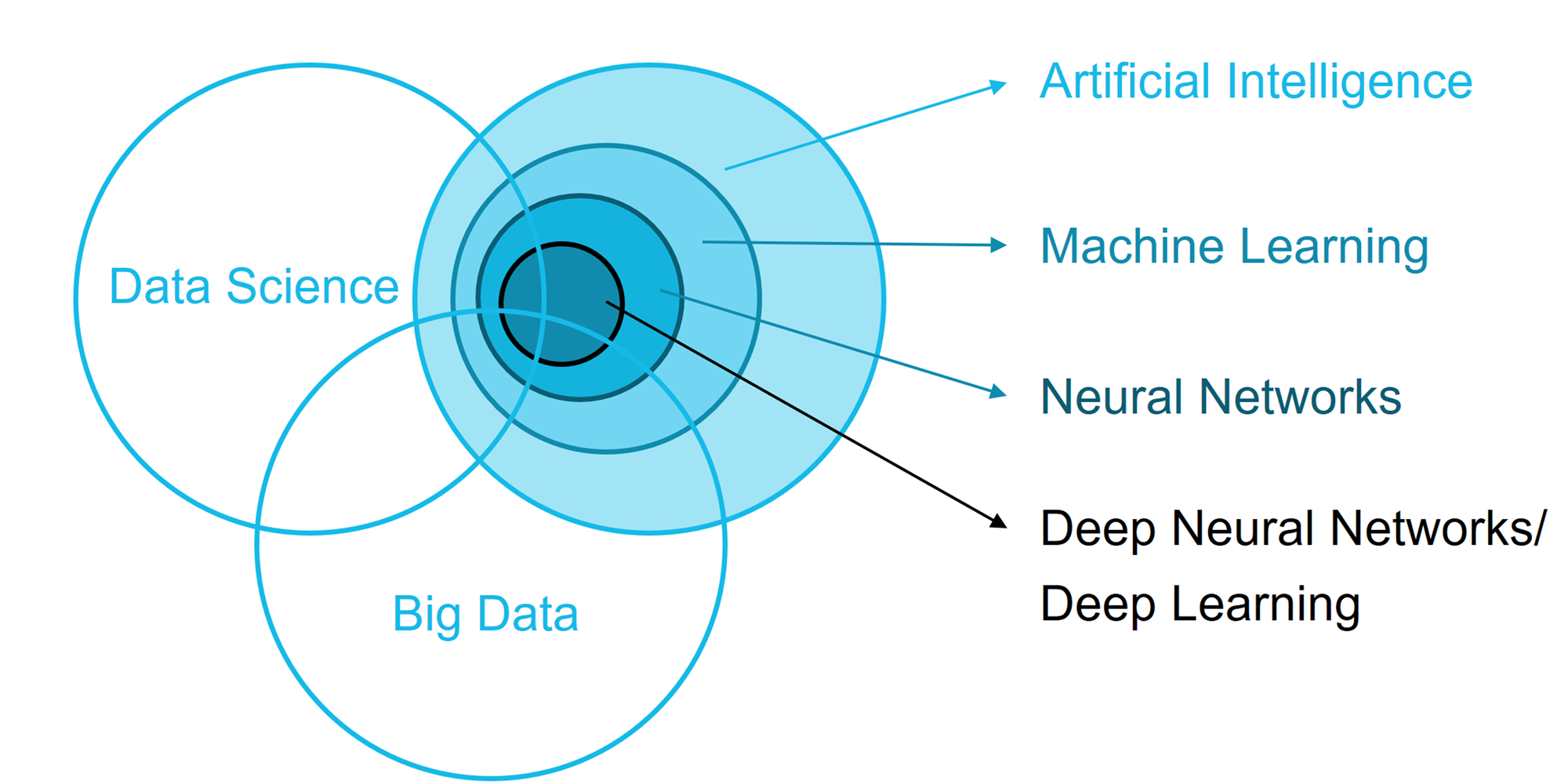
The prediction model is built with
top-notch statistical models
using artificial intelligence,
big data, and machine learning
approaches.
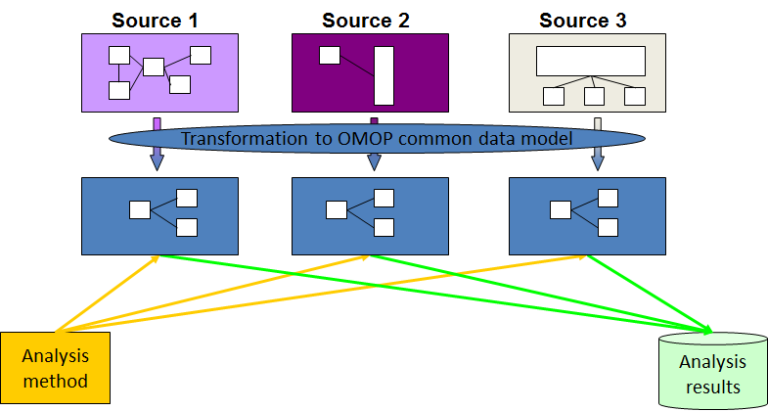
A kind of RWD, the Observational Medical Outcomes Partnership of the Common Data Model
(OMOP-CDM),
is a product of
harmonizing data collected for a different purpose (i.e., Electronic Medical Records (EMR) and
administrative claims
data).
OMOP-CDM consists of standardized data and information, including terminologies,
vocabularies, and coding schemes.
OMOP-CDM allows researchers to minimize information loss using standardized formats and perform
systematic analyses
using standardized analytic routines.
Research Questions
Main idea: Uric acid and its related metabolites in the purine metabolism pathway are early biomarkers for renal injury and proteinuria in non-diabetic CKD.Q1. Are SUA and its metabolites (esp. Allantoin) associated with CKD and ESRD incidence?
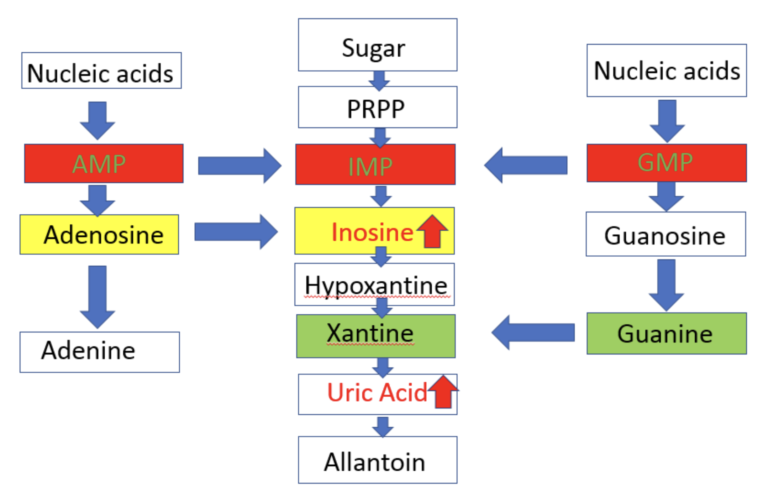
In collaboration with Drs. Cheryl Winkler, Jeffery Kopp, and Thorkell Andersson, we did
a pilot
study of untargeted
metabolite profiling in group (5 each) by HIVAN (HIV associated nephropathy) vs. HIV (+) control and
APOL1 high FSGS
(Focal Segmental Glomerulosclerosis) vs. APOL1 high risk healthy control.
Inosine and uric acid in purine pathway showed a significant increase.
Analysis Plan
- HIVAN selected from banked plasma collected as part of NIH cohort study
- Mann-Whitney U test for the comparison of the peaks: Corrected by using the false discovery rates (FDR)
- Logistic regression model to evaluate association of metabolite peak areas with the presence of HIVAN or CKD
- Identifying the significant peak areas, using the National Institute of Science and Technology (NIST) library and Metlin database if necessary
- After identifying significant metabolites, pathway analysis will be performed using Ingenuity Pathway Analysis (IPA)
- Kruskal-Wallis tests (non-parametric ANOVA) to identify differences in plasma metabolite concentrations between groups.
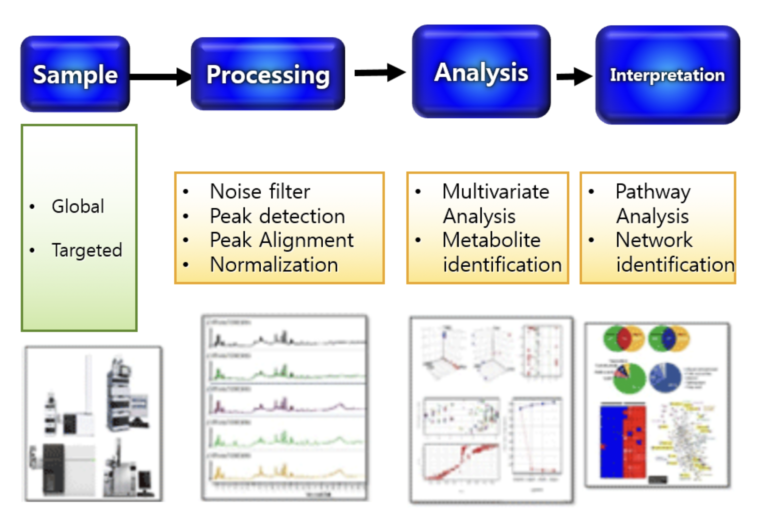
Q2. Is the genetic component of SUA related to CKD progression?
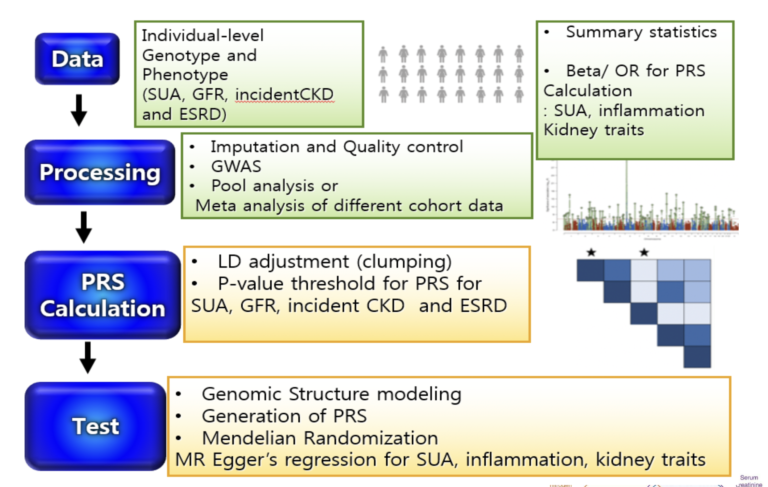
Analysis Plan
- GWAS of SUA and kidney trait (eGFR based on serum creatinine and on Cystatin C) in AAs for healthy and CKD cohorts
- Cross-trait LD Score regression (LDSR) to estimate genetic correlations between complex traits
- Genomic structural equation modeling (SEM) analyzes the joint genomic framework of complex phenotypes using GWAS summary statistics of individual traits with unknown overlap
- Polygenic risk score (PRS) to test for associations using the individual-level data of AAs from CRIC and AASK
- Two-sample MR analysis GWAS summary statistics for SUA, inflammation and kidney traits
to
test for putative causal
influence of
- SUA on kidney traits
- SUA on inflammation
- inflammation on kidney traits
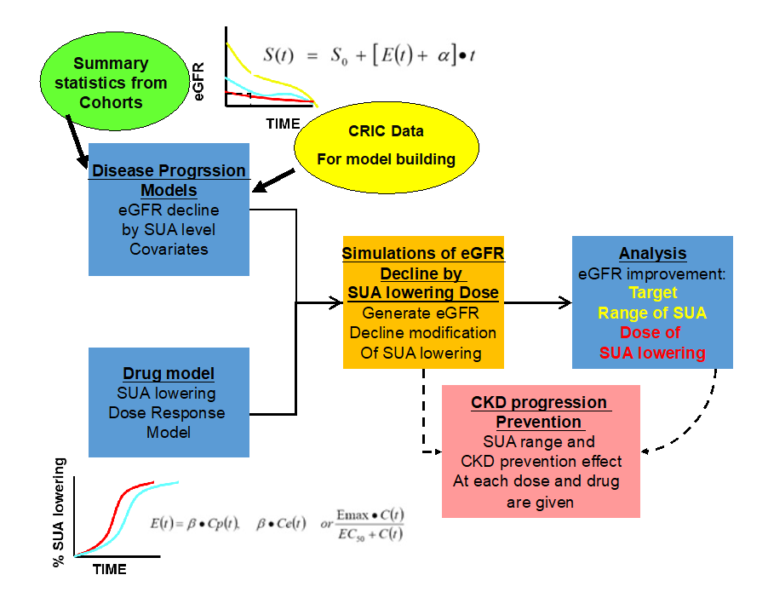
Q3. Does SUA modify factors in the non-diabetic CKD progression (by using the disease progression model)?
Analysis Plan
- Using the selected metabolites in Q1 & 2, build a disease progression model for non-diabetic CKD progression over time (esp. SUA)
- Build the drug model
- Based on the previous clinical trial results
- Linear, Emax, sigmoid Emax model and indirect response model
- Evaluation of how much lowering of SUA will benefit to CKD progression and suggest the dose range of each drug considering each individual disease status
Q4. Is the kidney function attributed to SUA?
Analysis Plan
- Using the population attributable fraction (PAF) we estimate the relative risk of the uric acid-related risk factors contributing to the incidence of hyperuricemia. Such estimation would be investigated across a different rage of the eGFR levels.
- We would be able to capture the amount of the relative risk of the uric acid-related risk factors contributing to the hyperuricemia, varied by the kidney functioning.
- The amount of the relative risk would be modulated by gender and ethnicity.
References
- Murphy, D., et al., Trends in Prevalence of Chronic Kidney Disease in the United States. Annals of Internal Medicine, 2016. 165(7): p. 473-481.
- Levin, A., et al., Global kidney health 2017 and beyond: a roadmap for closing gaps in care, research, and policy. Lancet, 2017. 390(10105): p. 1888-1917.
- Mwasongwe, S.E., et al., Relation of uric acid level to rapid kidney function decline and development of kidney disease: The Jackson Heart Study. Journal of Clinical Hypertension, 2018. 20(4): p. 775-783.
- Zoccali, C., et al., Uric acid and endothelial dysfunction in essential hypertension. Journal of the American Society of Nephrology, 2006. 17(5): p. 1466-1471.
- Zhou, Y., et al., Uric acid induces renal inflammation via activating tubular NF-κB signaling pathway. PLoS One, 2012. 7(6): p. e39738-e39738.
- Kurts, C., A crystal-clear mechanism of chronic kidney disease. Kidney international, 2013. 84(5): p. 859-861.
- Corry, D.B., et al., Uric acid stimulates vascular smooth muscle cell proliferation and oxidative stress via the vascular renin-angiotensin system. Journal of hypertension, 2008. 26(2): p. 269-275.
- Yu, M.A., et al., Oxidative stress with an activation of the renin-angiotensin system in human vascular endothelial cells as a novel mechanism of uric acid-induced endothelial dysfunction. Journal of hypertension, 2010. 28(6): p. 1234-1242.
- Netea, M.G., et al., The role of hyperuricemia in the increased cytokine production after lipopolysaccharide challenge in neutropenic mice. Blood, 1997. 89(2): p. 577-582.
- Kang, D., et al., A role for uric acid in the progression of renal disease. Journal of the American Society of Nephrology, 2002. 13(12): p. 2888-2897.
- Bellomo, G., et al., Association of uric acid with change in kidney function in healthy normotensive individuals. American journal of kidney diseases, 2010. 56(2): p. 264-272.
- Harambat, J., et al., Hyperuricemia after liver transplantation in children. Pediatric Transplantation, 2008. 12(8): p. 847-853.
- Cho, S.K., et al., U-Shaped Association Between Serum Uric Acid Level and Risk of Mortality: A Cohort Study. Arthritis Rheumatol, 2018. 70(7): p. 1122-1132.
- Srivastava, A., et al., Uric Acid and the Risks of Kidney Failure and Death in Individuals With CKD. American journal of kidney diseases, 2018. 71(3): p. 362-370.